40 how to predict splitting patterns
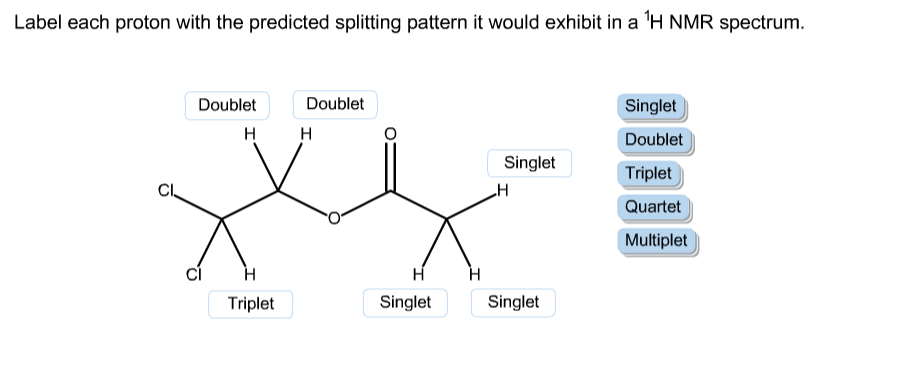
Names – GOV.UK Design System It’s also hard to predict the range of titles your users will have. If you have to ask for someone’s title, use an optional text input not a select. Remember to correctly use people’s names in any resulting correspondence. Allow users to change their name Answered: Predict the splitting pattern(s) you… | bartleby Science Chemistry Q&A Library Predict the splitting pattern(s) you would expect for the proton(s) in the molecule below that will show in its NMR spectrum. Explain all answers. CH; CH;CHCHCH; CH3.
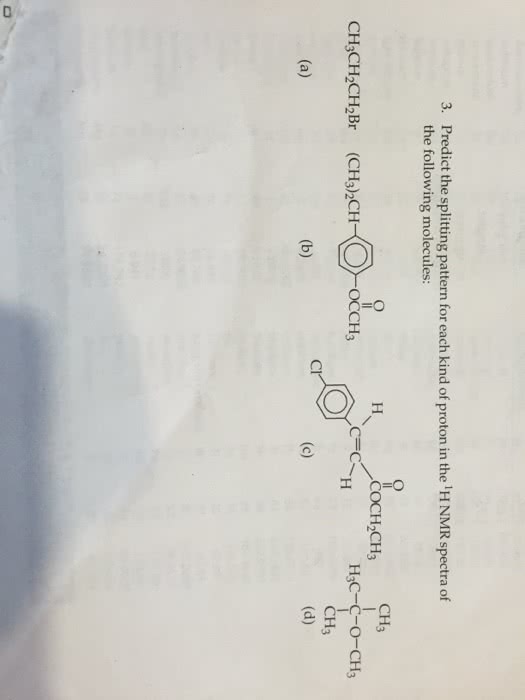
OneClass: Predict the splitting pattern for each kind of proton in ... Predict the splitting pattern for each kind of proton in the 1HNMRspectra of. the following molecules: (a) CH3CH2CH2Br (b)(CH3)2CHC6OCCH3 (c) ClC6(CH)2COCH2CH3 (d) (CH3)3COCH3. compou ent Carbon and protons are prese n Cacn ns nds, and thus predict the splitting patterns for certain proton (12 points). H3 H3 No. of nonequivalent carbon No. of ...

How to predict splitting patterns
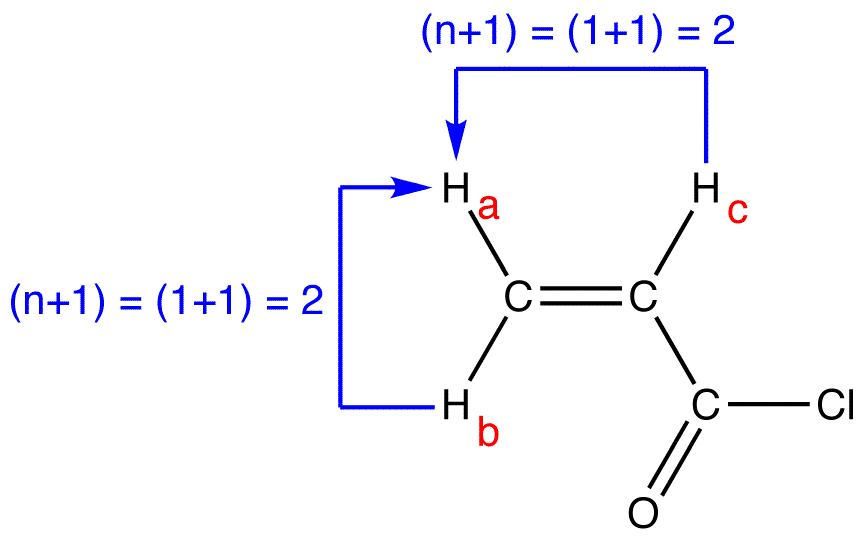
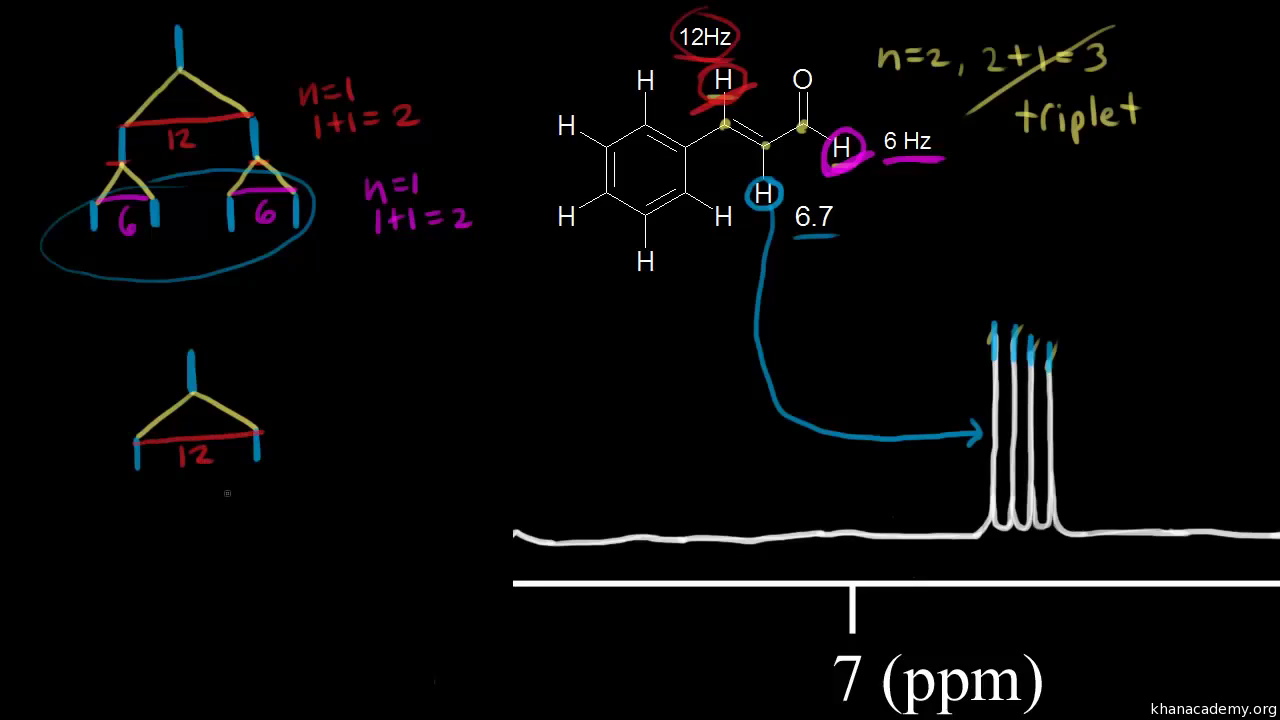
Multiplicity: n + 1 rule (video) | Khan Academy So one plus one is equal to two. We expect to see two peaks for the signal for the red proton. So here's the signal and we see our two peaks. So this is called a doublet. So the signal for the red proton is split in to two peaks because of the presence of the neighboring blue proton. Alright, let's do the same thing for the blue proton. Splitting and Multiplicity (N+1 rule) in NMR Spectroscopy - Chemistry Steps The more general formula for this is 2nI + 1, where I is the magnetic spin number of the given nucleus. And since it is equal to 1/2 for hydrogen, the formula that we use in 1 H NMR is n + 1. Below is a summary table for the splitting patterns in NMR spectroscopy. When two protons split each other's NMR signals, they are said to be coupled. Solved a 8. (8) Predict the splitting pattern for each | Chegg.com (8) Predict the splitting pattern for each signal in the 'H-NMR spectrum for the following molecule (singlet, doublet, triplet, quartet, pentet, sextet, etc). Enter your answer in the appropriate blank علیزم (a)_blank 1_ blank 2 (b) - (c) blank 3 (d) _____ blank 4 - (e) (f) blank 5_ blank 6 (g)______ blank 7___ (h)- blank 8
How to predict splitting patterns. Training, Validating and Testing - Towards Data Science Data Splitting. Single model case: In order to test our model with regard to its predictive accuracy it seems quite intuitive to split data into a training portion and a test portion, so that the model can be trained on one dataset, but tested on a different, new data portion. The reason for this test is simple, imagine we used the full dataset to train the model and then use the same data to ... NMR Signal Splitting N+1 Rule Multiplicity Practice Problems ... Using the splitting patterns shown above and the chemical shifts regions, predict the expected position and assign each peak to a group of protons (or one proton) based on the multiplicity of the signal. answer This content is for registered users only. Click here to Register! Making Predictions with Regression Analysis - Statistics By Jim The general procedure for using regression to make good predictions is the following: Research the subject-area so you can build on the work of others. This research helps with the subsequent steps. Collect data for the relevant variables. Specify and assess your regression model. Multiplet simulator - cheminfo Multiplet simulator. This tool allows to explain the shape of a signal as a function of its scalar couplings constants. It does not consider secondary effects. To start, you must define a set of coupling constants in the left upper table, along with its multiplicities. Coupling constants are specfied in Hz and multiplicities must be one of the ...
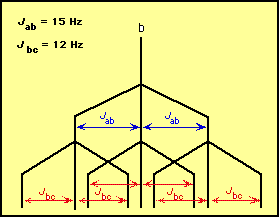
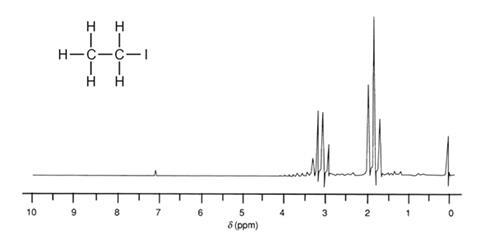
BASINS Framework and Features | US EPA May 06, 2022 · BASINS enables users to efficiently access nationwide environmental databases and local user-specified datasets, apply assessment and planning tools, and run a variety of proven nonpoint loading and water quality models within a single GIS format. How can I determine NMR splitting pattern? + Example - Socratic.org Nov 5, 2014 To find the NMR splitting pattern, for a given hydrogen atom, count how many identical hydrogen atoms are adjacent, and then add one to that number. For example, in CH 2ClCH 3 below, the red hydrogen atoms are adjacent to three identical hydrogen atoms (marked in blue). They will exhibit a quartet (4 peak; 3+1) splitting pattern. American policy is splitting, state by state, into two blocs Sep 02, 2022 · “I would predict that the effect is going to be that more and more red states are going to become more red, purple states are going to become red and the blue states are going to get a lot bluer,” Hawley said. “And I would look for Republicans as a result of this to extend their strength in the Electoral College. And that’s very good ... Complex splitting (video) | Proton NMR | Khan Academy So each line is split into a doublet. For this line and for this line. This line is split into a doublet, too. So we get four lines for the signal of the blue proton. If we go over here we see those four lines, one, two, three and four. And we call this a doublet of doublets, or a double doublet.
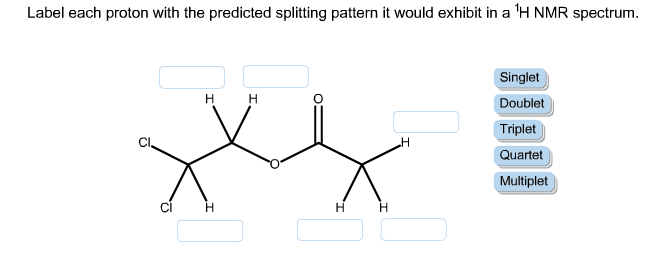
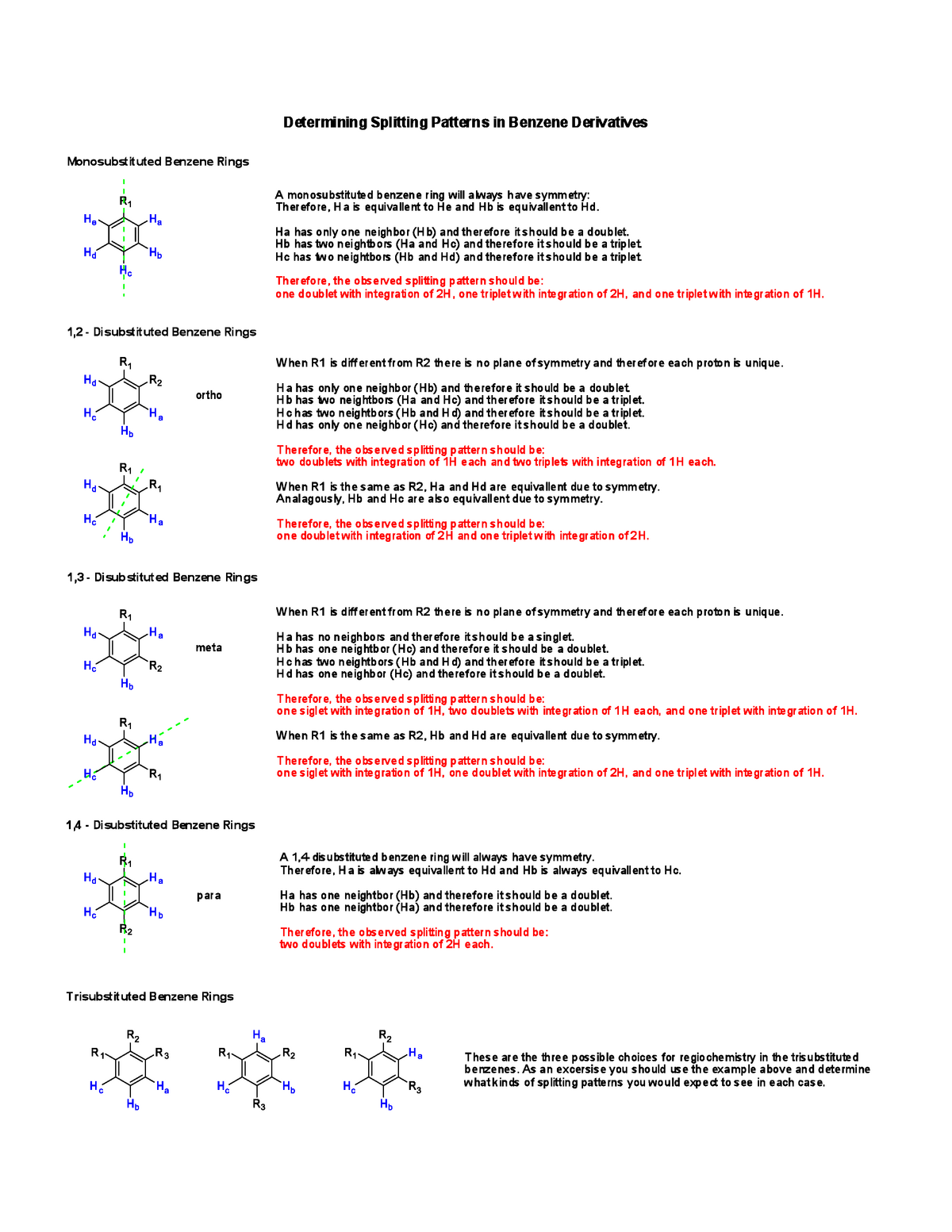
Assigning 1H NMR spectrum and predicting peak coupling patterns for 2H ... To predict the splitting patterns you anticipate for the aromatic system, consider first the general couplings you'd expect: ortho coupling ~7-8 Hz meta coupling ~2-3 Hz para coupling ~1Hz or less Let's ignore the para couplings for simplicity - they are not often seen. So, 16:10 Spin-Spin Splitting in ¹H NMR Spectra - Chemistry LibreTexts Predict the splitting pattern for the 1 H-NMR signals corresponding to the protons at the locations indicated by arrows (the structure is that of the neurotransmitter serotonin). Solutions Coupling constants Chemists quantify the spin-spin coupling effect using something called the coupling constant, which is abbreviated with the capital letter J. Learning Patterns - Speaker Deck Dec 20, 2021 · 2. Code Splitting: To avoid a large bundle of JavaScript code, you could start splitting your bundles. Code-Splitting is supported by bundlers like Webpack where it can be used to create multiple bundles that can be dynamically loaded at runtime. Code splitting also enables you to lazy load JavaScript resources. 3. Solved How can I easily predict the splitting patterns? How | Chegg.com Expert Answer. Transcribed image text: -> Predict Splitting Pattern -> Give Simplest ratio of the integration value for every # present Ha Hal Ho ho Hy Splitting Pattern Integration HO Hc Hdite Ota Нь Ha Pattern integration Hé Hc Hd Hc OH for the i rige plant HH Ho Hc c HO CS Scanned with CamScanner ce Hf Ha Hb He Hc Hal Hol Hel Hd He He Hd ...
How to predict the correct splitting pattern in a NMR spectrum my guess would be to compare the chemical shift difference between two nuclei, i and j with their coupling constant and when δ σ i j > j i j then i need to split the peaks corresponding to particle i and j but this would mean that coupling between nuclei that are very far apart with a very small j would also be split, which is not really what you …
How to calculate the splitting pattern for a proton NMR - YouTube About Press Copyright Contact us Creators Advertise Developers Terms Privacy Policy & Safety How YouTube works Test new features Press Copyright Contact us Creators ...
N + 1 Rule and Predicting Proton NMR Splitting (Multiplicity) DO NOT FORGET TO SUBSRCIBE!This video puts emphasis on proton NMR splitting patterns.
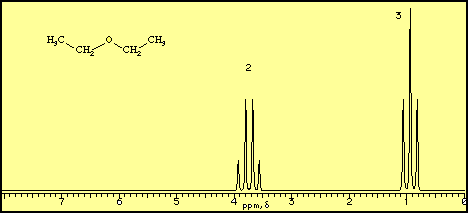
1H NMR - Splitting - Wake Forest University Predict the splitting patterns for the labeled hydrogens by drawing the peaks, represented by lines. Drag to draw the lines. Double-click on the line the delete it. (Hint: also note the height ratio of the peak and the distance between peaks.) For ethyl acetate, 1. Draw the splitting pattern for the hydrogens labeled A. 2.
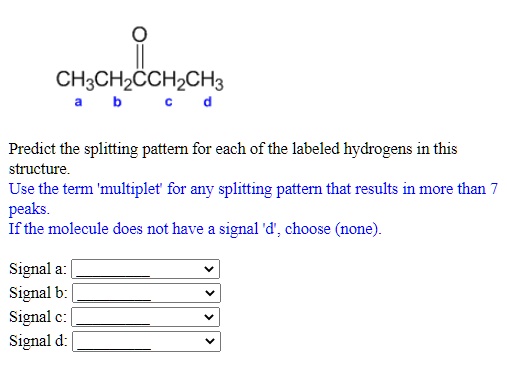
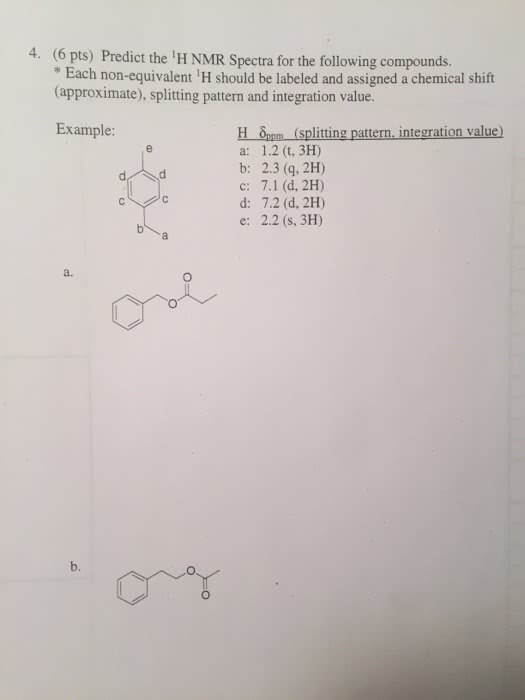
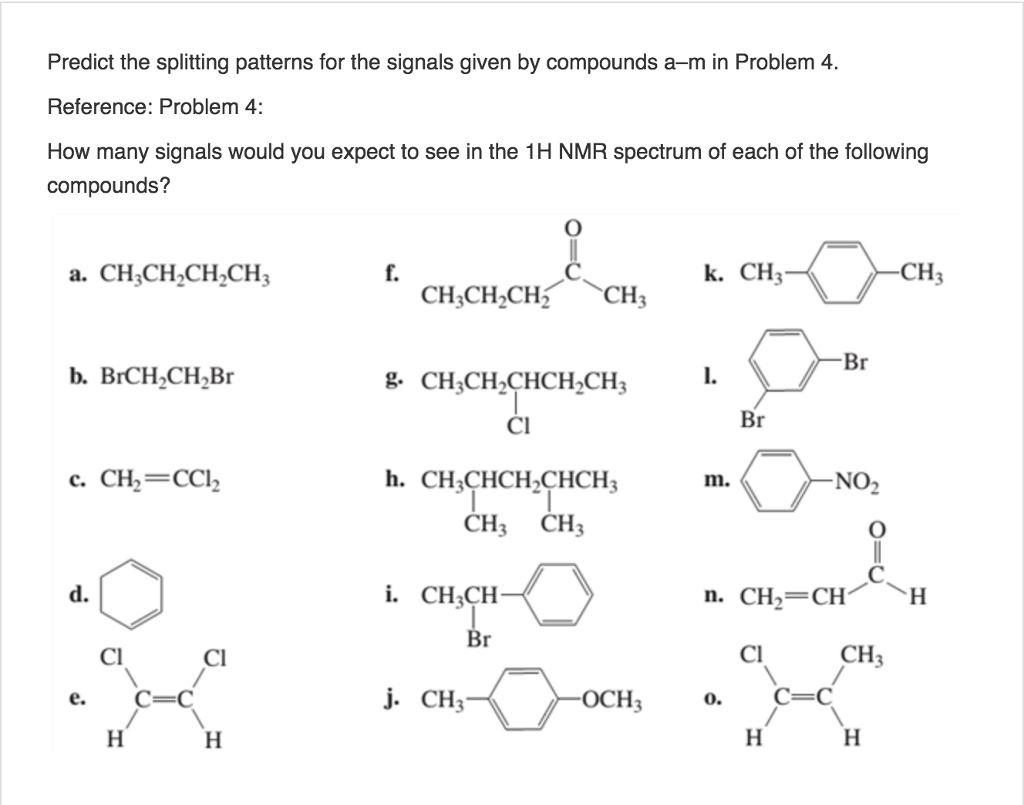
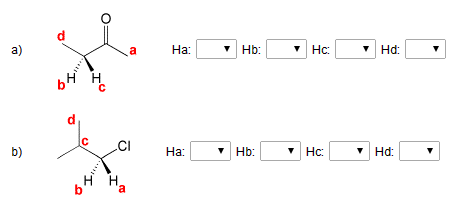
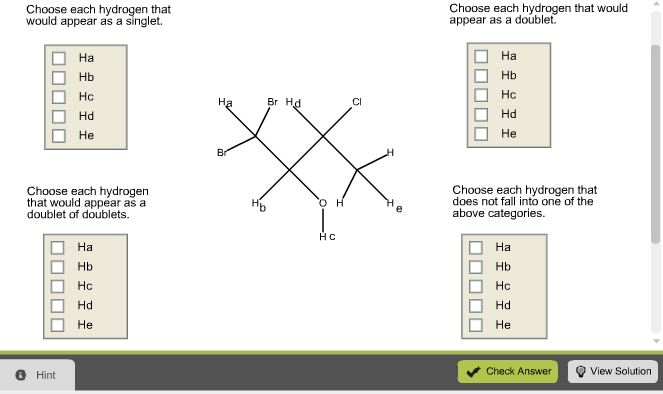
Predict the splitting pattern for each of the labeled hydrogens in the ... Predict the splitting pattern for each of the labeled hydrogens in the following molecules. Assume that all coupling constants are equal. a) Ha: Hb: Hc: Hd: b) Ha: 1 ... Considering the first question the rule used for prediction of splitting pattern is n+1 (Pascal's Triangle rule), where n is number of H atom on the adjacent carbon which are ...
Classification in Machine Learning: What it is and ... Aug 23, 2022 · Here a model can predict the existence of many known things in a photo, such as “person”, “apple”, "bicycle," etc. A particular photo may have multiple objects in the scene. This greatly contrasts with multi-class classification and binary classification, which anticipate a single class label for each occurrence.
PDF Predicting spin-spin coupling patterns in NMR spectra split each line of the doublet into a triplet (2x2x½ + 1 = 3). The CH2F protons therefore give rise to a doublet of triplets in the spectrum. The CH2 protons will be coupled to the CH2F protons, the F nucleus, and the CH3 protons. The peak will be split into 4 by the CH3 protons (2x3x½ + 1 = 4); each of these four peaks will be split into two
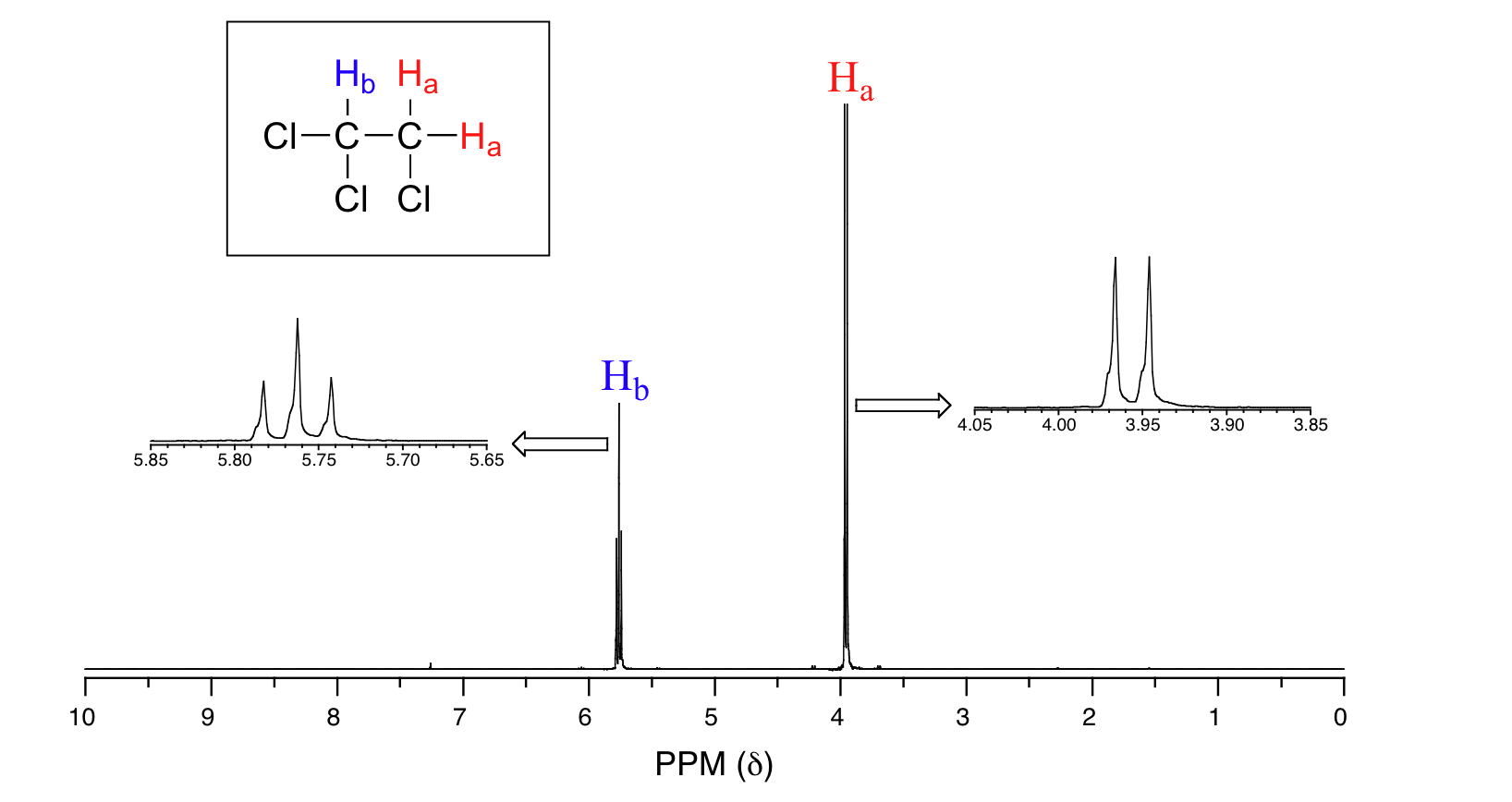
14.13: Splitting Diagrams Explain the Multiplicity of a Signal In this and in many spectra to follow, we show enlargements of individual signals so that the signal splitting patterns are recognizable. The signal at 3.96 ppm, corresponding to the two H a protons, is split into two subpeaks of equal height (and area) - this is referred to as a doublet. The H b signal at 5.76 ppm, on the other hand, is ...
Decision Tree Split Methods | Decision Tree Machine Learning Here are the steps to split a decision tree using reduction in variance: For each split, individually calculate the variance of each child node Calculate the variance of each split as the weighted average variance of child nodes Select the split with the lowest variance Perform steps 1-3 until completely homogeneous nodes are achieved
13.7: More Complex Spin-Spin Splitting Patterns Objectives. After completing this section, you should be able to. explain how multiple coupling can give rise to complex-looking 1 H NMR spectra.; predict the splitting pattern expected in the 1 H NMR spectrum of an organic compound in which multiple coupling is possible.; interpret 1 H NMR spectra in which multiple coupling is evident.
How can I draw NMR splitting diagrams? | Socratic The general rule for ""^1"H" NMR spectra is: n neighbouring protons with the same coupling constant J will split a signal into n+1 lines. We draw a "splitting tree" for each proton or group of protons. No Adjacent Protons No splitting. Draw a single line. 1 Adjacent Proton Draw 2 lines (a doublet) separated by J_(ax). 2 Adjacent Protons From each line of the doublet, draw two more lines ...
Spin-spin splitting and coupling - More complex splitting patterns More complex splitting patterns 1 H NMR patterns are more complex than predicted by the N+1 coupling rule when coupling of one proton or set of equivalent protons occurs to two different sets of protons with different size coupling constants or when coupling occurs between protons with similar but not identical chemical shifts.
How would you create a system? to predict patterns to predict patterns in pick3 and pick4. The bjective is instead of predicting digits. or numbers is to predict positional patterns. to the number is or is not in the pattern. from 0, 4 to 5, 9 or ...
Train/Test Split and Cross Validation - A Python Tutorial Enter the validation set. From now on we will split our training data into two sets. We will keep the majority of the data for training, but separate out a small fraction to reserve for validation. A good rule of thumb is to use something around an 70:30 to 80:20 training:validation split.
mass spectra - fragmentation patterns - chemguide The m/z = 29 peak is produced by the ethyl ion - which once again could be formed by splitting the molecular ion either side of the CO group. Peak heights and the stability of ions. The more stable an ion is, the more likely it is to form. The more of a particular sort of ion that's formed, the higher its peak height will be.
Continuous Delivery for Machine Learning - Martin Fowler Sep 19, 2019 · This usually involves splitting the data into a training set and a validation set, trying different combinations of algorithms, and tuning their parameters and hyper-parameters. That produces a model that can be evaluated against the validation set, to assess the quality of its predictions.
Solved a 8. (8) Predict the splitting pattern for each | Chegg.com (8) Predict the splitting pattern for each signal in the 'H-NMR spectrum for the following molecule (singlet, doublet, triplet, quartet, pentet, sextet, etc). Enter your answer in the appropriate blank علیزم (a)_blank 1_ blank 2 (b) - (c) blank 3 (d) _____ blank 4 - (e) (f) blank 5_ blank 6 (g)______ blank 7___ (h)- blank 8
Splitting and Multiplicity (N+1 rule) in NMR Spectroscopy - Chemistry Steps The more general formula for this is 2nI + 1, where I is the magnetic spin number of the given nucleus. And since it is equal to 1/2 for hydrogen, the formula that we use in 1 H NMR is n + 1. Below is a summary table for the splitting patterns in NMR spectroscopy. When two protons split each other's NMR signals, they are said to be coupled.
Multiplicity: n + 1 rule (video) | Khan Academy So one plus one is equal to two. We expect to see two peaks for the signal for the red proton. So here's the signal and we see our two peaks. So this is called a doublet. So the signal for the red proton is split in to two peaks because of the presence of the neighboring blue proton. Alright, let's do the same thing for the blue proton.








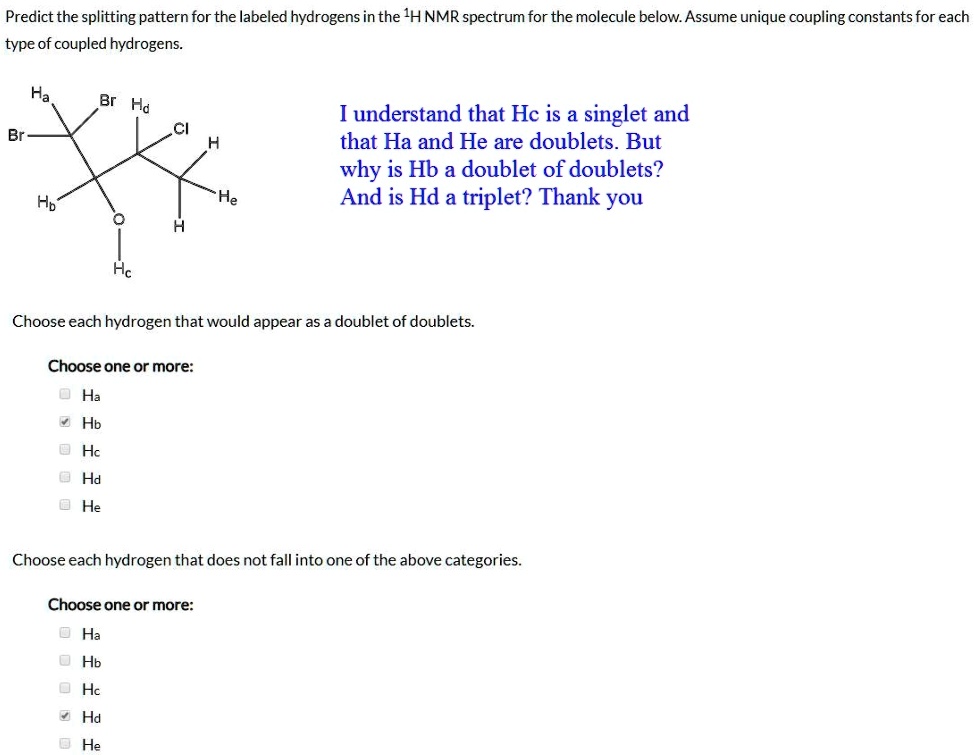








Post a Comment for "40 how to predict splitting patterns"